Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
石田 恒
Proteins: Structure, Function, and Bioinformatics, 82(9), p.1985 - 1999, 2014/09
被引用回数:10 パーセンタイル:32.21(Biochemistry & Molecular Biology)Proteasome is involved in the degradation of proteins. Proteasome activators bind to the proteasome core particle (CP) and facilitate opening a gate of the CP, where Tyr8 and Asp9 in the N-termini tails of the CP form the ordered open gate. Four different molecular dynamics simulations were carried out: ordered- and Tyr8Gly/Asp9Gly disordered-gate models of the CP complexed with an ATP-independent PA26 and ordered- and disordered-gate models of the CP complexed with an ATP-dependent PAN-like activator. In the ordered-gate models, the substrate in the activator was more stable than that in the CP. In the disordered-gate models, the substrate in the activator was more destabilized than in the ordered-gate models. Thus, it was concluded that the dynamics of the N-termini tails entropically play a key role in the translocation of the substrate.
Poornam, G. P.*; 松本 淳; 石田 恒; Hayward, S.*
Proteins: Structure, Function, and Bioinformatics, 76(1), p.201 - 212, 2008/12
被引用回数:83 パーセンタイル:91.05(Biochemistry & Molecular Biology)A new method for the analysis of domain movements in large, multichain, biomolecular complexes is presented. The method is applicable to any molecule for which two atomic structures are available that represent a conformational change indicating a possible domain movement. The method is blind to atomic bonding and atom type and can, therefore, be applied to biomolecular complexes containing different constituent molecules such as protein, RNA, or DNA. Here, we report on the application of the method to biomolecules covering a considerable size range: hemoglobin, liver alcohol dehydrogenase, S-Adenosylhomocysteine hydrolase, aspartate transcarbamylase, and the 70S ribosome. The results provide a depiction of the conformational change within each molecule that is easily understood, giving a perspective that is expected to lead to new insights.
茶竹 俊行; Ostermann, A.; 栗原 和男; Parak, F.*; 新村 信雄
Proteins: Structure, Function, and Bioinformatics, 50(3), p.516 - 523, 2003/02
被引用回数:54 パーセンタイル:76.7(Biochemistry & Molecular Biology)蛋白質や核酸などの生体分子は三次元構築して初めてその機能を発現する。その構造構築の過程において、水素原子や水和水は重要な役割を果たしているが、これらの立体構造を決定できる方法は中性子結晶解析のみである。本研究では、三種類の蛋白質について1.5~1.6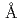 分解能での中性子結晶構造解析を行い、水和構造の詳細を明らかにすることに成功した。中性子核密度図上で水分子はさまざまな形をとっており、しかも、この形状と水分子のダイナミクスとの間に重要な相関が見いだされた。今回明らかになった水分子のダイナミクスは、コンピューターシミュレーションやNMRでの溶媒和解析の研究に対する有益な情報となることが希望される。
分解能での中性子結晶構造解析を行い、水和構造の詳細を明らかにすることに成功した。中性子核密度図上で水分子はさまざまな形をとっており、しかも、この形状と水分子のダイナミクスとの間に重要な相関が見いだされた。今回明らかになった水分子のダイナミクスは、コンピューターシミュレーションやNMRでの溶媒和解析の研究に対する有益な情報となることが希望される。